Google Summer of Code
Hi! Welcome to my personal notebook on my GSoC 2023 journey. In this summer I worked with Dr. Augustin Luna and Dr. Bo Yuan on converting CellBox, a model predicting cell pertubation effects under various drug combinations, from Tensorflow to Pytorch. This notebook serves as my weekly goals record. For some more detailed comments I have during my coding period, please check out my Google Docs personal notebook. This is my final Pytorch implementation of CellBox.
Overview
CellBox (original repo and paper) is a deep learning model that predicts perturbation effects on cells when applied with different combinations of drugs. It takes in as input a matrix of normalized combinations of drug doses (shape=(89, 89), corresponding to 89 unique drug combinations), and it predicts expressions of different proteins within the cell for each drug combination (shape=(89, 99), corresponding to 99 protein + phenotypic nodes for all 89 drug combinations). 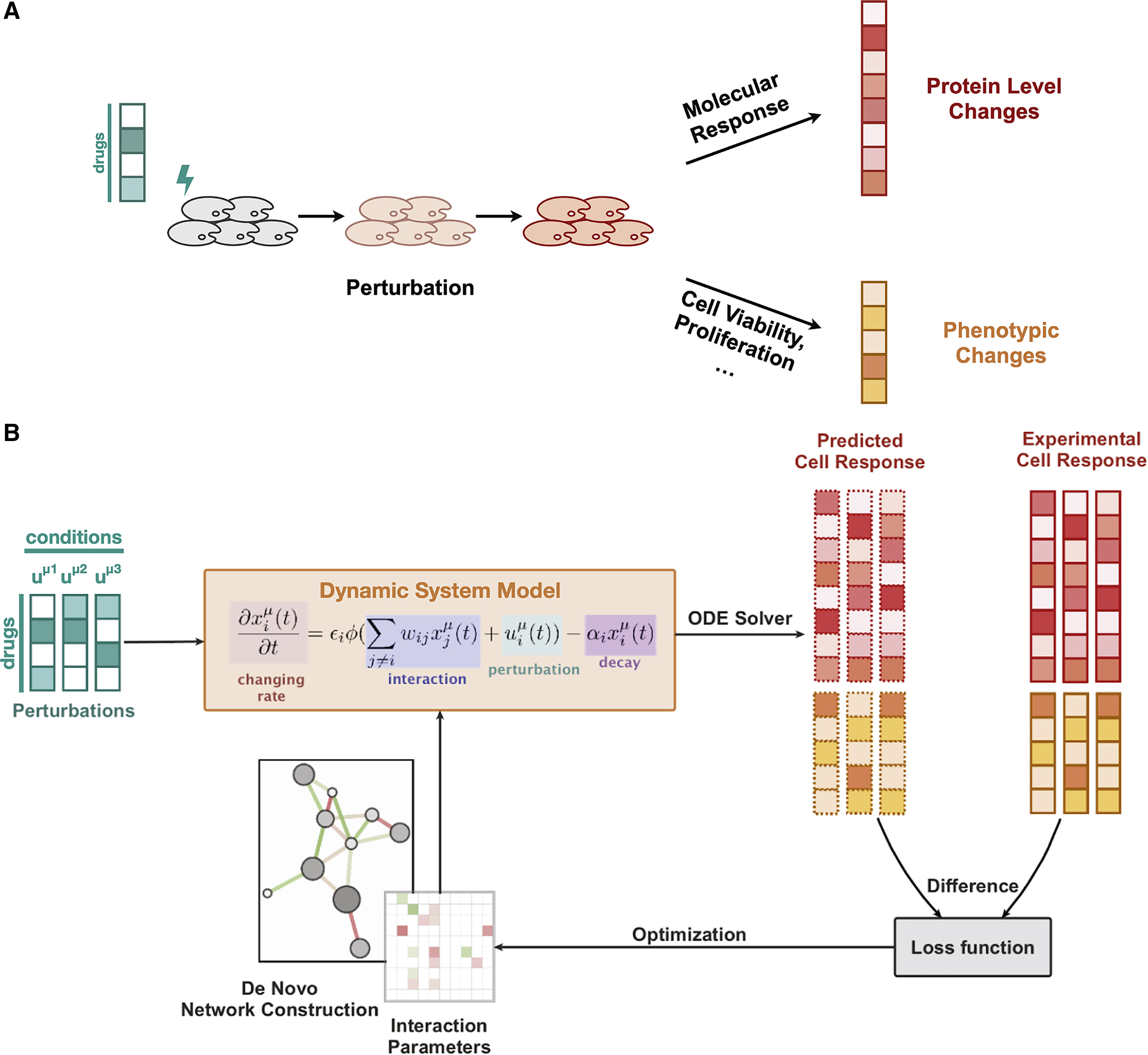
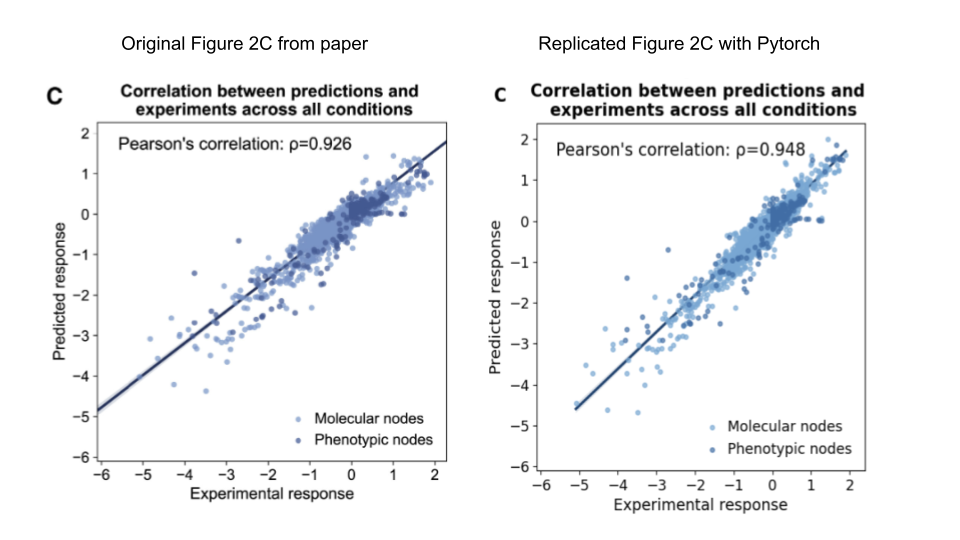
The original CellBox implementation is in Tensorflow 1 and includes many deprecated functions. Therefore, this project aims to convert it to Pytorch so that other researchers are more familiar with the codebase. The resulting repo completely removes Tensorflow from the codebase. This repo passes extensive tests that ensure similar performance compared with the original repo, the most significant result being a full replication of Figure 2C in the original paper. This figure was created by training 500 models with different random seeds and taking the average prediction of all 500 models.